

Non-canonical quadruplexes (G4) in the nucleic acids represents specific secondary structures that correlated and participate important biological process, including telomeric propagation, tumor cell proliferation and are close to the life span. The interaction of G4 with specific proteins and monitoring the unfolding process is important to understand the development and evolution of some diseases and for further regulation of telomere and disclose the mechanism of typical cancers.
Recently, the Chongqing Institute of Green and Intelligent Technology, Chinese Academy of Sciences (CAS), has achieved fruitful results in nanopore technology research. The related findings have been published in the academic journal Nucleic Acids Research.
In the study of nucleic acid-protein interactions, the research team employed label-free nanopore single-molecule technology to achieve, for the first time, the direct real-time monitoring of the unwinding process of DNA/RNA quadruplexes (G-quadruplexes, G4) by specific proteins (TEP1, RTEL1, nsp13) (Fig. 1). The study successfully observed that a 20 nM human telomeric DNA G4 sequence (hTel) in a 0.5 M CsCl buffer at weakly acidic conditions (pH 5) could be efficiently unwound into single-stranded DNA (ssDNA) within 1 hour by 10 nM TEP1 or RTEL1 protein. Notably, the TEP1 protein exhibited selective unwinding capability for hybrid-topology G4 structures, while showing minimal activity towards parallel G4s (Fig. 2).
In research on the SARS-CoV-2 virus RNA, the team confirmed that RNA G4 structures derived from the viral genome (e.g., RNA-1574) could be efficiently unwound by its helicase nsp13 under conditions of 2 M LiCl and pH 5. Significant unwinding was achieved within 1 hour using only a 50% molar ratio of nsp13 relative to the G4 structure. This technology provides a novel single-molecule research tool for understanding the mechanistic roles of G4 structures in key biological processes such as telomere maintenance, tumorigenesis, and viral replication. (Reference: Unwinding Process of DNA/RNA Quadruplexes by Proteins under Label-Free Nanopore Monitoring, Nucleic Acids Res.)
Related paper links: DOI: 10.1093/nar/gkaf54
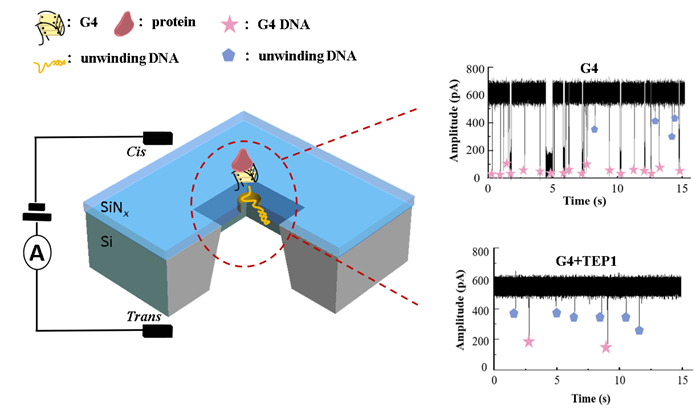
Fig. 1. Unwinding recording of quadruplexes via nanopore sensors
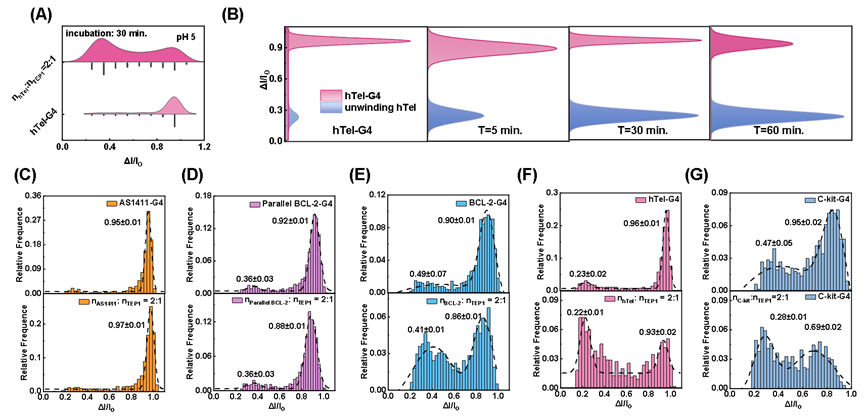
Fig. 2. Unfolding selectivity of TEP1 with G4 of distinct topologies.