

The tethered state of biomolecules is a key focus in studies such as single-molecule physics and single-molecule sequencing. As an intermediate state, a tethered molecule exhibits some free diffusion properties at one end, while the other end is strictly restricted by a molecular anchor, resembling a fixed state. However, the electrodynamics involved in molecular diffusion, dielectrophoresis, and stretching processes of tethered molecules remain unclear. Studying tethered states is scientifically significant for revealing the motion models and molecular properties of such molecules in solution.

ACS Nano Cover, October 2014 Issue
Recently, the Chongqing Institute of Green and Intelligent Technology, Chinese Academy of Sciences, in collaboration with Wuhan University, Changchun University of Science and Technology, and the University of Oulu in Finland, published a paper in ACS Nano titled “Directly Characterizing the Capture Radius of Tethered Double-stranded DNA by Single-Molecule Nanopipette Manipulation” The paper was selected as the cover article for the October 2014 issue of the journal. In this research, the team developed a Single-molecule Manipulation, Identification, and Length Examination (SMILE) system based on nanopipettes. Using this system, they successfully captured and stretched tethered double-stranded DNA molecules. The results demonstrated characteristic capture (rcapture) and stretch (rstretch) radii surrounding the DNA anchor point.
Notably, the study observed a consistent ratio of characteristic capture radius across different DNA lengths at varying capture voltages, closely matching their corresponding gyration radius ratio. For the stretch radius, the ratio was consistent with the contour length (L0), and the stretch ratio (rstretch/L0) increased from 70% to 90% as the voltage increased (100 mV to 1000 mV). More importantly, through numerical simulations, the team identified that the characteristic capture and stretch radii were influenced by entropy elasticity and the electric field, respectively, which act as capture and escape barriers. This research introduces a new single-molecule manipulation and analysis method, which holds promise for comprehensive exploration of the conformation, electrical, and diffusion properties of tethered molecules.
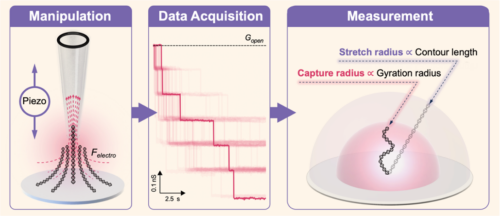
SMILE Single-Molecule Manipulation System
This study is the first to reveal the characteristic length distribution of tethered molecules captured by nanopores. Several findings indicate that the gyration radius of tethered molecules approximates the capture radius under low-voltage conditions, providing both experimental and theoretical foundations for uncovering the inherent physical properties of tethered molecules. Additionally, this research introduces innovative measurement methods and perspectives for fields such as biophysics, gene sequencing, and molecular diagnostics involving charged molecules like RNA and proteins.
This work was supported by the National Key R&D Program, the National Natural Science Foundation of China, the Youth Science Fund, and the Natural Science Foundation of Chongqing.
Link to the related paper: https://doi.org/10.1021/acsnano.4c05605